Juyoun YooI am currently a postdoc in Franck Polleux lab in Zuckerman Institute at Columbia University. I did my PhD training with Larry Zipursky via Neuroscience IDP program at UCLA.
My research interest is in how the brain is built. Thesedays I'm fascinated by how the gene regulatory programs are temporally coordinated during brain development. |

|
ResearchWork during my Ph.D in Zipursky lab |
|
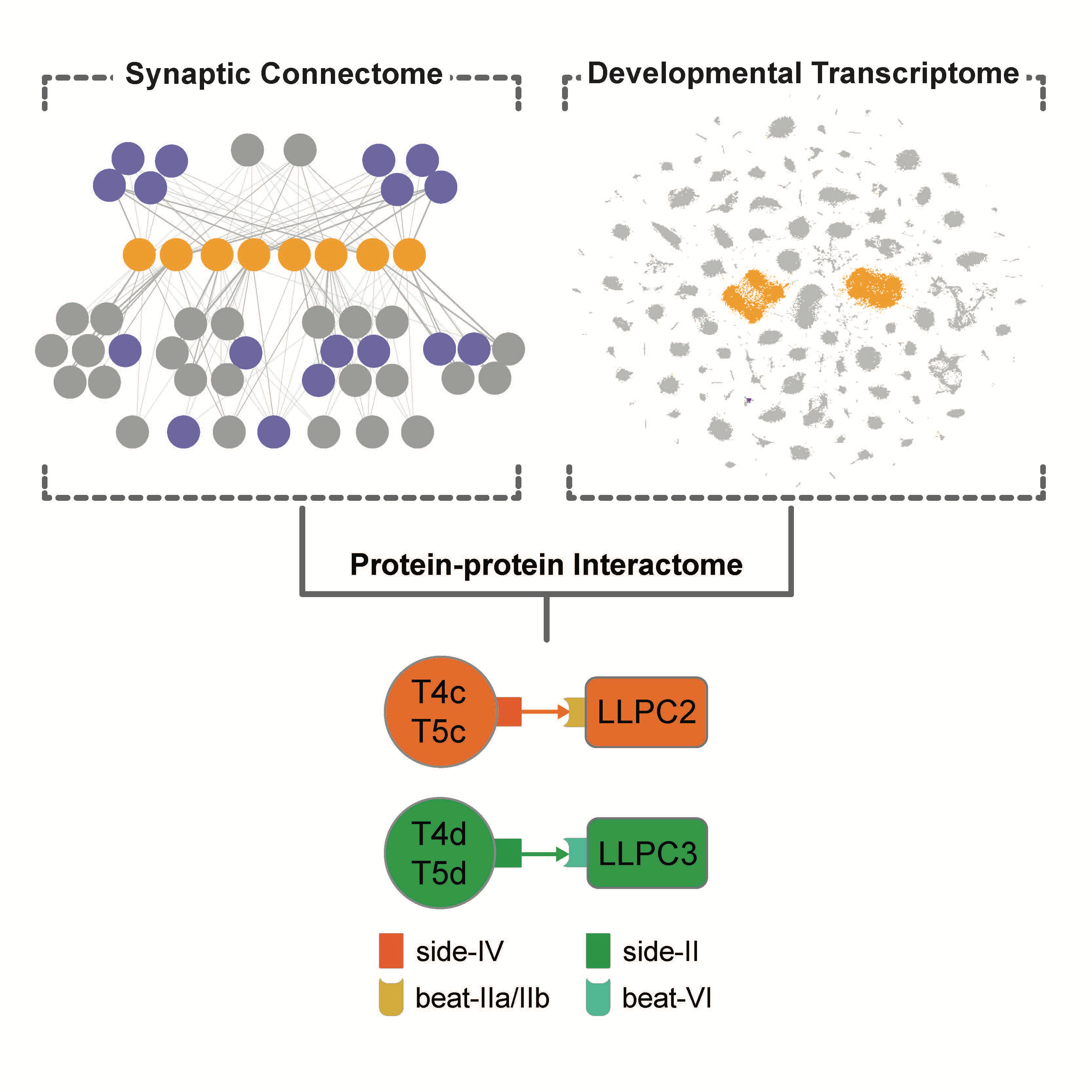
Transcriptional Programs of Circuit Assembly in the Drosophila Visual SystemJuyoun Yoo, Mark Dombrovski, Parmis Mirshahidi, Aljoscha Nern, Samuel A. LoCascio, S. Lawrence Zipursky, Yerbol Z. KurmangaliyevCurr Biol, 2023. [ We mapped the developemental transcriptome we generated in 2020 and the electron microscopic connectome from Janelia. We identified matching expression of IgSF proteins Beat and Side in the parallel synaptic partners in the motion dectection circuit. These protein family binding specificity also matched with the synaptic partner choices. I generated CRISPR KO allels for beat/side genes and demonstrated that these receptor-ligand pairs are required for optimal synaptic partner decisions. |
|
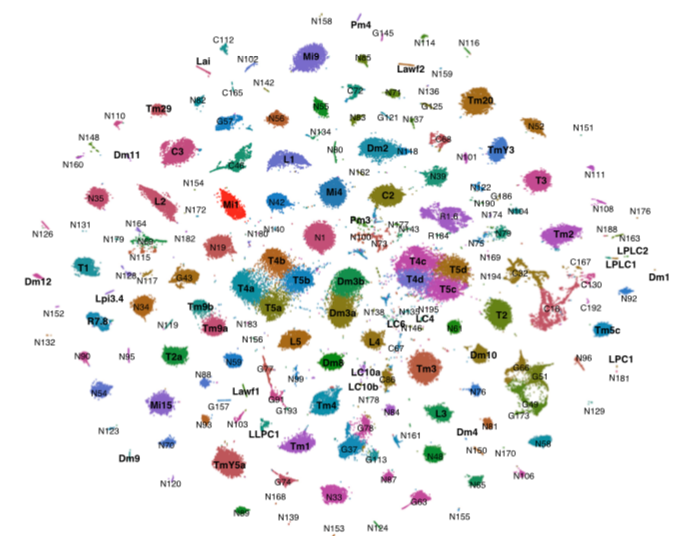
Transcriptional Programs of Circuit Assembly in the Drosophila Visual SystemYerbol Z. Kurmangaliyev, Juyoun Yoo, Javier Valdes-Aleman, Piero Sanfilippo, S. Lawrence ZipurskyNeuron, 2020. [ We generated developmental transcriptome atlas of the whole fly opticlobe throughout the pupal stage. We showed that there are pan-neuronal programs that the majority of neurons follow, and there also are cell-type specific programs. We strategized to use Drosophila Genome Reference Panel(DGRP) to multiplex the library to minimize batch effect in a simple and cost-effective manner. |
|
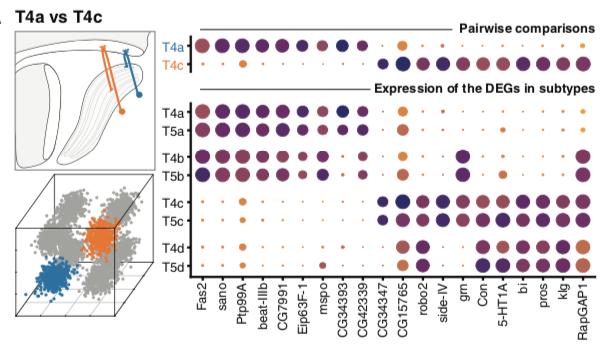
Modular transcriptional programs separately define axon and dendrite connectivityYerbol Z Kurmangaliyev*, Juyoun Yoo*, Samuel A LoCascio*, S Lawrence ZipurskyeLife, 2019. [ We showed that eight subtypes of direction selective T4/T5 neurons' transcriptome has no subtype-specific markers, rather the architecture of the transcriptome is assembled with axon and dendrite specific programs, reflecting the morphologies. |
|
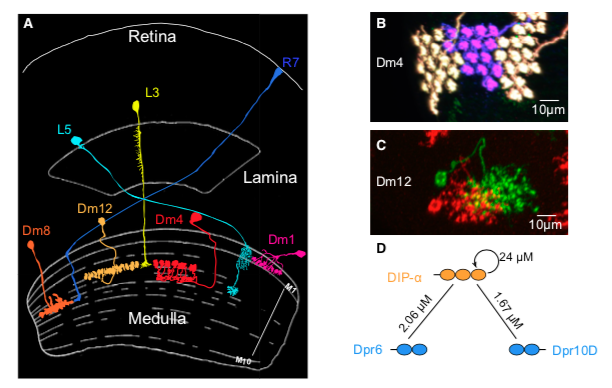
Interactions between the Ig-Superfamily Proteins DIP-α and Dpr6/10 Regulate Assembly of Neural CircuitsShuwa Xu, Qi Xiao, Filip Cosmanescu, Alina P. Sergeeva, Juyoun Yoo, Ying Lin, Phinikoula S. Katsamba, Goran Ahlsen, Jonathan Kaufman, Nikhil T. Linaval, Pei-Tseng Lee, Hugo J. Bellen, Lawrence Shapiro, Barry Honig, Liming Tan, and S. Lawrence ZipurskyNeuron, 2018. [ We showed that Immunoglobulin Superfamily proteins DIP-α and its binding partners dpr6, dpr10 interactions regulate layer specific circuit assembly. |